Proxima
Reprograming protein-protein interactions to unlock new small-molecule therapies for cancer, autoimmune disorders, and other hard-to-treat diseases
Proteins, carriers of the signals that can spark or stop cellular functions, seldom work alone — they’re found in complexes with other proteins. Among drug hunters there’s been a growing hope that if we could understand, control, and disrupt how proteins lock together, it might give us new ways to slow difficult-to-treat diseases like cancer or autoimmune disorders.
There’s been good reason for this hope, as a number of “molecular glue” molecules such as Revolution Medicines’ daraxonrasib, which slips between two proteins called cyclophilin A and RAS to block cancer signaling, have advanced in clinical trials. What has limited progress is that there’s been no systematic way to identify or design molecular glues: biologists tend to stumble across them serendipitously, one at a time.
There’s one player, Proxima, working to change that. Today Bloomberg broke the news that the company (formerly known as VantAI) has raised $80 million in seed-stage financing to double down on its AI-driven technology for the rational discovery and design of new molecular glues and other proximity-based, small-molecule medicines. DCVC led the round, joined by NVIDIA NVentures (our partner at another TechBio portfolio company, Relation, in London), Magnetic Ventures (also our partner at Relation), and other investors.
For DCVC, it’s the largest TechBio investment to date for any investment through Series A. Proxima has already secured multibillion-dollar research collaborations with big biopharma companies, and we think their work is important because it bears out — in new and exciting ways — our driving hypothesis about TechBio: namely, that state-of-the-art AI combined with high-value, proprietary biological data can not only uncover new medicines, but make the whole drug discovery process faster, cheaper, and more scalable.
The idea of molecular glues and other kinds of “proximity modulators” is not new. PROTACs or proteolysis targeting chimeras — pairs of binding substances called ligands that can eliminate unwanted proteins by binding on one end to a target protein and, on the other, to ligases that cause them to break down — were one early example. Dozens of PROTAC-based drug candidates are now in clinical trials, and last year Johnson & Johnson bought PROTACs maker Halda Therapeutics (a Proxima partner) for $3 billion. Daraxonrasib and other next-generation small-molecule proximity modulators are different from PROTACs because they’re usually single ligands, inspired by natural molecular glues such as rapamycin or cyclosporin, but with novel features and mechanisms of action that could allow researchers to program protein-protein interactions more precisely.
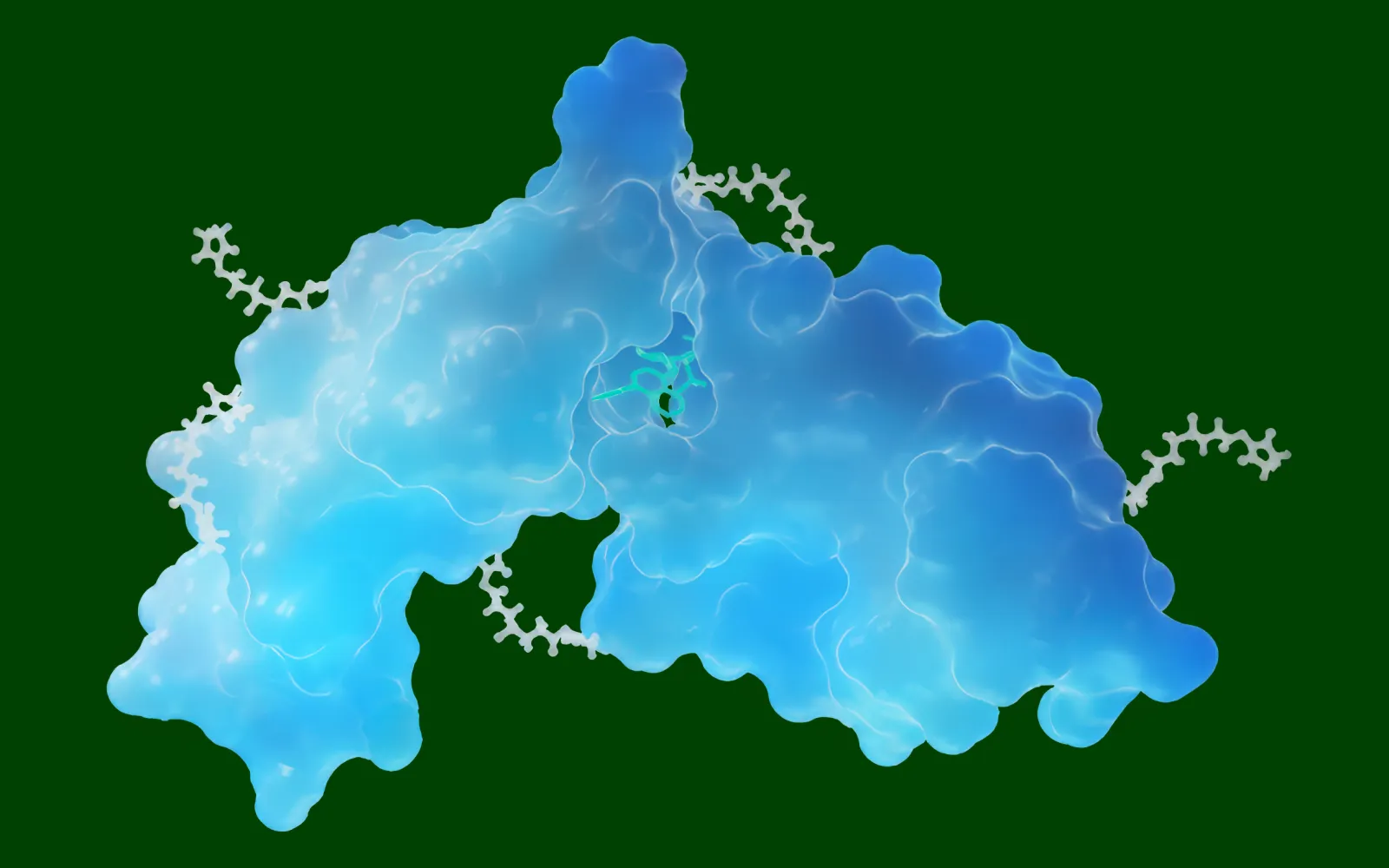
Proxima, founded by its CEO Zachary Carpenter and CTO Luca Naef, spun out from Roivant in 2023 with a classic TechBio agenda: to speed up discovery of new proximity modulators by, first, building a vast proprietary atlas of real-world biological interactions between proteins, and, secondly, using that data to train a generative machine learning model. In Proxima’s case, that model can simultaneously simulate both how proteins co-fold and design new ligands that might interface with these proteins and give them new functions.
Michael Bronstein, Proxima’s chief scientist-in-residence, is the DeepMind professor of AI at the University of Oxford and a pioneer in the field of geometric deep learning. His work formed the basis for Proxima’s Neo series of diffusion-based frontier models, which are built to analyze protein-protein and protein-ligand-protein interactions. Neo is trained on a growing database of cross-linking mass spectrometry (XLMS) data that includes copious examples of cross-linked peptides denatured and digested from native proteins or whole cells. (This database spans virtually the whole proteome, including most known enzymes and transcription factors.) Bronstein and Proxima’s insight was that this training would give Neo‑1 an efficient way to carry out generative design tasks in structural biology — like reasoning from the amino-acid sequences for two given proteins to a hypothesized structure for a small-molecule ligand that will fit into predicted pockets of both proteins and bind them, or inhibit binding.
Already, Proxima is applying this unique tech stack in business deals to help partners identify targets or effector molecules for their own drug discovery programs. Collaborations with Johnson & Johnson, Bristol Myers Squibb, and Blueprint Medicines (now part of Sanofi) have the potential to bring in billions of dollars in upfront and milestone payments and royalties. And that’s just the beginning. As Proxima builds up its team of scientists, physicians, and programmers, it’ll be well positioned to grow an internal pipeline of novel treatments for complex diseases.
We see Proxima as part of a crucial reform movement in biotechnology, one that’s using frontier AI and proprietary lab data to find more and better drug targets, faster. We owe it to patients to accelerate progress on polygenic conditions such as cancer, autoimmune disease, and neurodegeneration, where there’s often been little or no material advance in decades. When it’s easier to find novel targets and molecules to cure disease, we can test many more of them — which not only yields more hits but lowers the cost of failure. We’ve invested across multiple companies implementing this TechBio philosophy in unique ways, from Recursion Pharmaceuticals to Relation, Noetik, and Totus. We think Proxima has a huge data and computational advantage on a frontier — proximity-based therapeutics — that could yield molecules that hit multiple undruggable targets and intractable diseases. That’s why we were so proud to lead the investment unveiled this week.